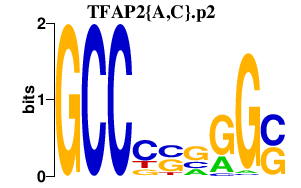
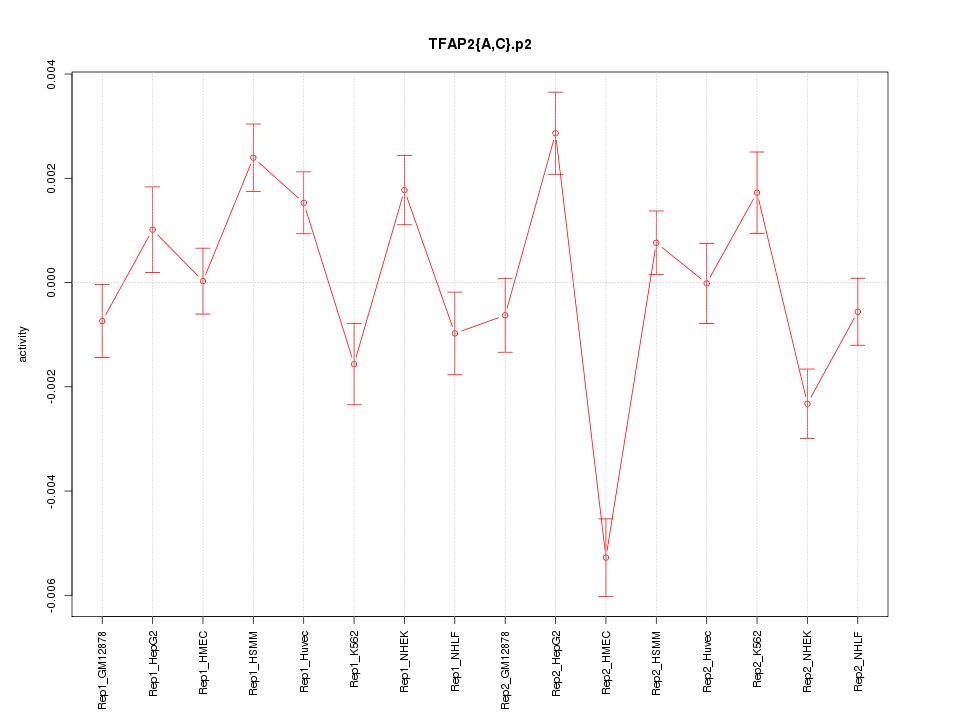
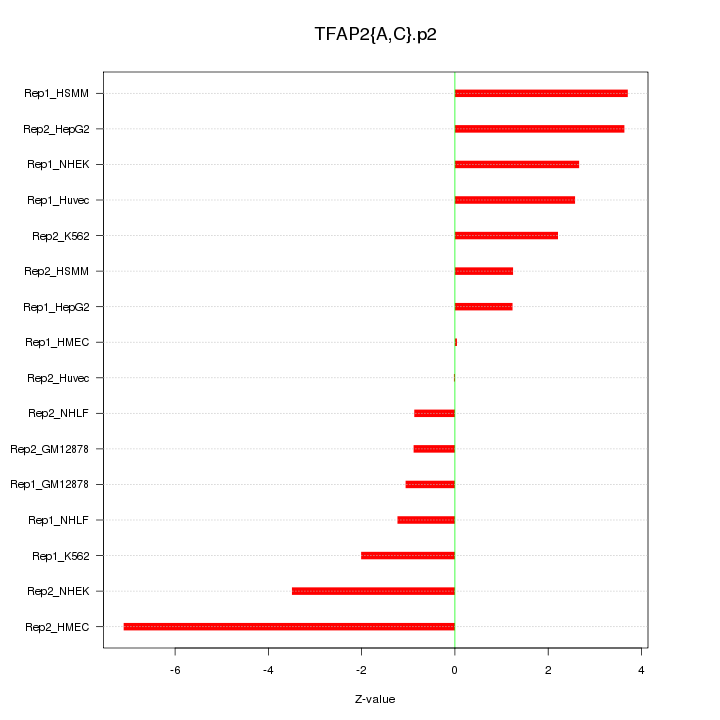
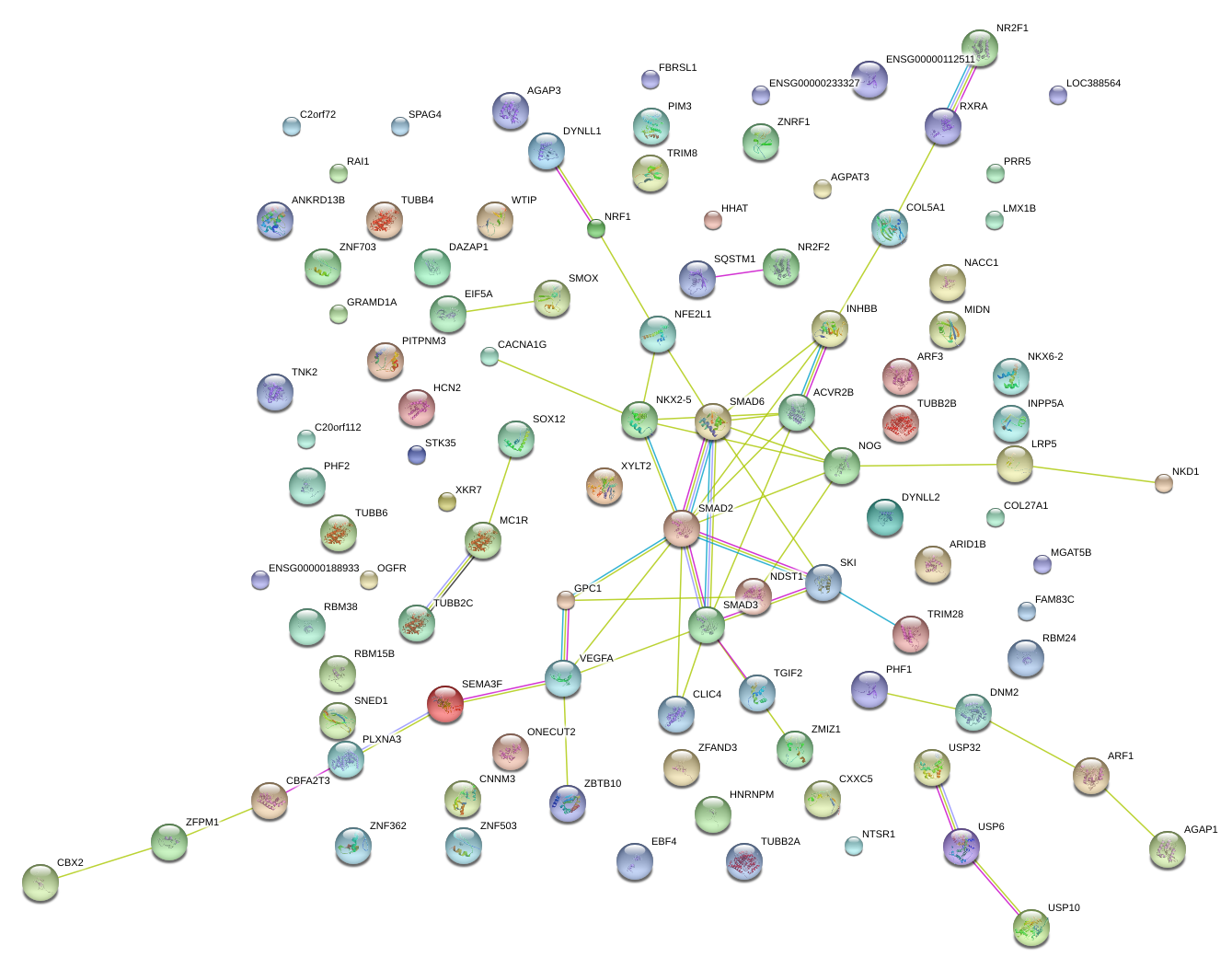
|
chr10_+_104394185
|
6.114
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr2_+_236068029
|
4.040
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr2_+_96845704
|
3.182
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr19_+_540849
|
2.898
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr20_+_254205
|
2.785
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr2_+_236067687
|
2.670
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr20_+_2031418
|
2.659
|
|
STK35
|
serine/threonine kinase 35
|
|
chr7_+_150414704
|
2.569
|
NM_001042535
NM_031946
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr1_+_2149993
|
2.531
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr20_+_34635362
|
2.529
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr6_+_157141565
|
2.465
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr20_-_30636535
|
2.404
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr17_+_72376022
|
2.379
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr9_+_128416542
|
2.211
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr10_+_80672842
|
2.166
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr19_+_1200882
|
2.164
|
|
MIDN
|
midnolin
|
|
chr3_+_38470782
|
2.163
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr20_-_2031521
|
2.149
|
|
|
|
|
chr16_-_87535106
|
2.112
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr3_+_51403760
|
2.101
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr7_+_150442652
|
2.095
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr9_+_136673534
|
2.063
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr12_+_131577229
|
2.051
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr17_+_17525571
|
2.019
|
|
|
|
|
chr15_+_64782468
|
2.005
|
|
SMAD6
|
SMAD family member 6
|
|
chr16_+_88517187
|
1.977
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chr2_+_236067424
|
1.932
|
NM_001037131
NM_014914
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr20_+_60906605
|
1.898
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr9_+_136358145
|
1.803
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr6_+_37895570
|
1.786
|
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chrX_+_153339814
|
1.737
|
NM_017514
|
PLXNA3
|
plexin A3
|
|
chr17_+_17525497
|
1.729
|
NM_030665
|
RAI1
|
retinoic acid induced 1
|
|
chr5_+_139008667
|
1.721
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr2_+_241023908
|
1.701
|
|
GPC1
|
glypican 1
|
|
chr8_+_81560971
|
1.699
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr16_+_87047418
|
1.699
|
NM_153813
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr22_+_48740432
|
1.691
|
|
PIM3
|
pim-3 oncogene
|
|
chr16_+_49139694
|
1.613
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr15_+_94674849
|
1.612
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_+_50217692
|
1.597
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr5_+_179180695
|
1.593
|
|
SQSTM1
|
sequestosome 1
|
|
chr10_+_80498782
|
1.547
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr5_+_179180509
|
1.537
|
|
SQSTM1
|
sequestosome 1
|
|
chr16_+_83291112
|
1.537
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr8_+_37672427
|
1.534
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr19_+_63747829
|
1.510
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr6_+_37895244
|
1.506
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr1_+_33494745
|
1.502
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr19_+_10689813
|
1.499
|
|
DNM2
|
dynamin 2
|
|
chr20_-_34635091
|
1.480
|
|
|
|
|
chr9_+_95378492
|
1.459
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr17_+_45993338
|
1.457
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr6_+_157140954
|
1.448
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr20_+_4077466
|
1.442
|
|
SMOX
|
spermine oxidase
|
|
chr9_+_115958051
|
1.435
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr19_+_40183169
|
1.432
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr2_+_120820140
|
1.422
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr6_+_43846155
|
1.398
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_226337052
|
1.390
|
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr11_+_67836575
|
1.386
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr9_+_136358134
|
1.382
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr20_+_30019465
|
1.375
|
NM_001011718
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7
|
|
chr2_+_241023880
|
1.372
|
|
|
|
|
chr5_+_179180517
|
1.371
|
|
SQSTM1
|
sequestosome 1
|
|
chr20_+_2030527
|
1.364
|
NM_080836
|
STK35
|
serine/threonine kinase 35
|
|
chr9_+_136673464
|
1.360
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr17_+_75366524
|
1.346
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr19_+_1199548
|
1.341
|
NM_177401
|
MIDN
|
midnolin
|
|
chr15_+_65145215
|
1.337
|
|
SMAD3
|
SMAD family member 3
|
|
chr16_+_73590739
|
1.328
|
|
ZNRF1
|
zinc and ring finger 1
|
|
chr2_+_241586927
|
1.320
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr5_+_149845503
|
1.318
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr20_+_55399847
|
1.299
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr17_-_6400470
|
1.296
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr17_+_7152417
|
1.295
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr22_+_43476710
|
1.286
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr10_-_134449467
|
1.268
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr1_+_24944554
|
1.266
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr2_+_231610438
|
1.265
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr19_+_1358750
|
1.255
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr17_+_45778381
|
1.248
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr17_+_52026058
|
1.234
|
NM_005450
|
NOG
|
noggin
|
|
chr19_+_8415869
|
1.229
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr20_+_33667160
|
1.226
|
NM_003116
|
SPAG4
|
sperm associated antigen 4
|
|
chr9_+_115957537
|
1.215
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr17_+_53515725
|
1.212
|
NM_080677
|
DYNLL2
|
dynein, light chain, LC8-type 2
|
|
chr19_+_8415875
|
1.204
|
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr10_-_76829857
|
1.202
|
|
ZNF503
|
zinc finger protein 503
|
|
chr19_+_13090101
|
1.202
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr5_+_92946348
|
1.201
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr10_+_134201262
|
1.199
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr19_-_60587414
|
1.184
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr18_+_53253775
|
1.184
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr16_+_73590375
|
1.181
|
NM_032268
|
ZNRF1
|
zinc and ring finger 1
|
|
chr6_+_157140895
|
1.163
|
|
|
|
|
chr16_+_83291097
|
1.163
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr7_+_129038765
|
1.157
|
NM_005011
|
NRF1
|
nuclear respiratory factor 1
|
|
chr17_+_24944605
|
1.156
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr21_+_44109529
|
1.152
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr20_-_33343617
|
1.150
|
NM_178468
|
FAM83C
|
family with sequence similarity 83, member C
|
|
chr16_+_87047159
|
1.143
|
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr20_+_60810610
|
1.140
|
NM_002531
|
NTSR1
|
neurotensin receptor 1 (high affinity)
|
|
chr16_+_83291055
|
1.127
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr20_+_2621193
|
1.123
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr3_-_197120276
|
1.123
|
NM_005781
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr5_-_172594691
|
1.123
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr3_+_50167851
|
1.120
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr19_+_39664706
|
1.119
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr20_+_56899904
|
1.119
|
|
GNAS
|
GNAS complex locus
|
|
chr10_+_98582648
|
1.106
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr20_-_55274497
|
1.100
|
|
BMP7
|
bone morphogenetic protein 7
|
|
chr2_-_47650973
|
1.095
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr22_+_18091065
|
1.088
|
NM_000407
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide
|
|
chr2_+_238540411
|
1.085
|
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative)
|
|
chr3_+_185515543
|
1.084
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_-_120715937
|
1.083
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr4_+_3434830
|
1.076
|
NM_001164673
NM_173660
|
DOK7
|
docking protein 7
|
|
chr5_+_179180447
|
1.066
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr1_+_226336983
|
1.058
|
NM_001024226
NM_001658
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr15_+_72861767
|
1.053
|
|
CSK
|
c-src tyrosine kinase
|
|
chr19_+_3958690
|
1.053
|
NM_015897
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr22_+_43476755
|
1.052
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr2_+_241023703
|
1.046
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr20_+_226200
|
1.040
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr17_+_43480723
|
1.035
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr16_+_73590419
|
1.031
|
|
ZNRF1
|
zinc and ring finger 1
|
|
chr1_+_226337016
|
1.031
|
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr2_+_36436316
|
1.029
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr21_+_44109454
|
1.026
|
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr18_+_18969724
|
1.021
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr17_+_43480724
|
1.021
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr15_+_72861470
|
1.019
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr16_+_73590470
|
1.018
|
|
ZNRF1
|
zinc and ring finger 1
|
|
chr19_-_1799447
|
1.018
|
NM_020695
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)
|
|
chr2_+_36436873
|
1.017
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr19_+_18803786
|
1.014
|
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr17_-_76064842
|
1.012
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr22_+_36531246
|
1.008
|
|
H1F0
|
H1 histone family, member 0
|
|
chr16_+_85158357
|
1.008
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr6_+_43846372
|
1.007
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_9571518
|
1.005
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr5_+_36912656
|
1.004
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr16_-_695719
|
1.003
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr16_+_83291107
|
1.002
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr2_+_171280944
|
0.984
|
|
SP5
|
Sp5 transcription factor
|
|
chr16_+_88154638
|
0.983
|
|
RPL13
|
ribosomal protein L13
|
|
chr17_+_43480674
|
0.982
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr19_+_8415802
|
0.981
|
NM_005968
NM_031203
|
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr7_+_39956633
|
0.980
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr19_+_63747678
|
0.979
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr19_+_3136965
|
0.973
|
|
NCLN
|
nicalin
|
|
chr10_+_134060681
|
0.972
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr12_+_119456538
|
0.972
|
|
RNF10
|
ring finger protein 10
|
|
chr16_+_24648446
|
0.971
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr20_-_55275078
|
0.969
|
NM_001719
|
BMP7
|
bone morphogenetic protein 7
|
|
chr10_+_82204046
|
0.969
|
|
TSPAN14
|
tetraspanin 14
|
|
chr15_+_65145248
|
0.966
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr6_+_43846735
|
0.963
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr17_+_77780177
|
0.963
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr19_+_3136927
|
0.963
|
|
NCLN
|
nicalin
|
|
chr10_+_82204051
|
0.961
|
|
TSPAN14
|
tetraspanin 14
|
|
chr4_-_7039661
|
0.956
|
|
LOC100134937
|
hypothetical LOC100134937
|
|
chr19_+_3045528
|
0.955
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr1_+_224317062
|
0.953
|
|
H3F3A
LOC440926
|
H3 histone, family 3A
H3 histone, family 3A pseudogene
|
|
chr14_+_23633101
|
0.953
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr5_+_139008091
|
0.953
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr17_+_34140030
|
0.949
|
NM_001136498
|
CISD3
|
CDGSH iron sulfur domain 3
|
|
chr2_+_238540428
|
0.948
|
NM_080678
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative)
|
|
chr10_-_32676090
|
0.947
|
NM_025209
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr6_+_1556543
|
0.946
|
|
FOXC1
|
forkhead box C1
|
|
chr17_+_38186141
|
0.939
|
NM_032387
|
WNK4
|
WNK lysine deficient protein kinase 4
|
|
chr10_+_82204054
|
0.939
|
|
TSPAN14
|
tetraspanin 14
|
|
chrX_+_152606945
|
0.937
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr9_+_134026836
|
0.935
|
|
NTNG2
|
netrin G2
|
|
chr16_+_84204424
|
0.933
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr4_+_110574274
|
0.931
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr19_+_10689772
|
0.928
|
|
DNM2
|
dynamin 2
|
|
chr16_+_3447957
|
0.927
|
NM_001083600
NM_024845
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr19_+_7566698
|
0.926
|
NM_001080429
NM_020902
|
KIAA1543
|
KIAA1543
|
|
chr14_+_104226769
|
0.922
|
NM_001031714
NM_022489
NM_032714
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr19_+_63747638
|
0.922
|
NM_005762
|
TRIM28
|
tripartite motif containing 28
|
|
chr7_+_4688248
|
0.919
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr16_+_15950707
|
0.916
|
NM_004996
NM_019862
NM_019898
NM_019899
NM_019900
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr1_+_51474583
|
0.915
|
|
RNF11
|
ring finger protein 11
|
|
chr19_+_2220529
|
0.914
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr8_+_142471290
|
0.912
|
|
|
|
|
chr16_+_28741861
|
0.912
|
|
ATXN2L
|
ataxin 2-like
|
|
chr19_+_482737
|
0.911
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr19_+_10689763
|
0.909
|
|
DNM2
|
dynamin 2
|
|
chr20_+_29656744
|
0.907
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr1_+_24944493
|
0.904
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr1_-_25129305
|
0.903
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr6_+_37245860
|
0.902
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr6_+_43247179
|
0.900
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr19_+_2115119
|
0.899
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr7_+_288051
|
0.899
|
NM_020223
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr19_+_2220526
|
0.898
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr7_-_129038701
|
0.897
|
|
|
|
|
chr17_+_72880967
|
0.897
|
|
SEPT9
|
septin 9
|
|
chr19_+_4920126
|
0.896
|
|
KDM4B
|
lysine (K)-specific demethylase 4B
|